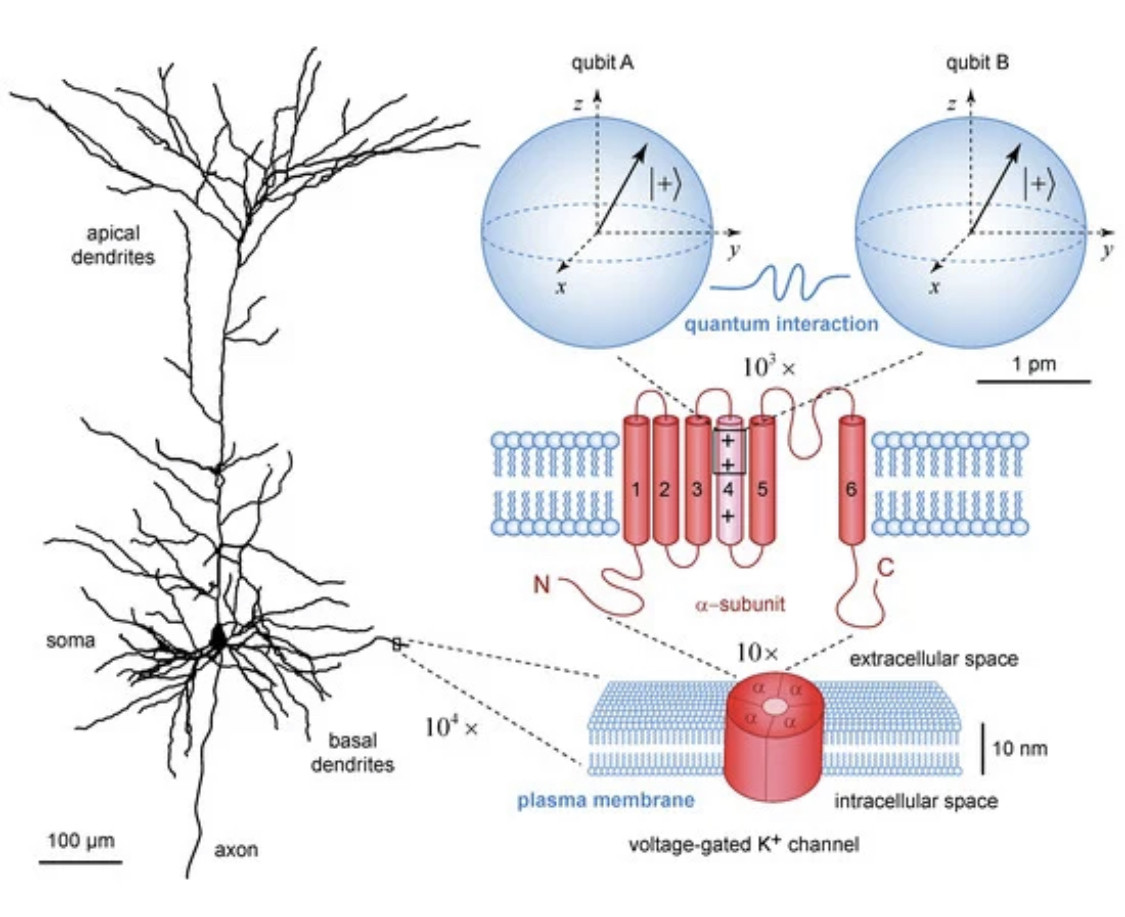
The presented results show that quantum coherence of individual subsystems cannot be used for cognitive binding because it is a physical mechanism that leads to separability and non-interaction. In contrast, quantum interactions with their associated decoherence of individual subsystems are instrumental for dynamical changes in the quantum entanglement of the composite quantum state vector and manifested correlations of different observable outcomes. Thus, fast decoherence timescales could assist cognitive binding through quantum entanglement across extensive neural networks in the brain cortex.
Mom, Granny, # Christian and #RedPilled for years . #GodBlessPresidentTrump #TheBestIsYetToCome
Trump 2020, Fight Like a Flynn ⭐️⭐️⭐️, WWG1WGA, Seth Rich, God Wins 🙏🏻, We Are The Storm 🦅, God Bless America 🇺🇸
Dear God... 🙏🏻🙏🏻🙏🏻
https://www.pnas.org/doi/10.1073/pnas.1119827109
Graphene is an ideal thin membrane substrate for creating molecule-scale devices. Here we demonstrate a scalable method for creating extremely small structures in graphene with atomic precision. It consists of inducing defect nucleation centers with energetic ions, followed by edge-selective electron recoil sputtering. As a first application, we create graphene nanopores with radii as small as 3 Å, which corresponds to 10 atoms removed. We observe carbon atom removal from the nanopore edge in situ using an aberration-corrected electron microscope, measure the cross-section for the process, and obtain a mean edge atom displacement energy of 14.1 ± 0.1 eV. This approach does not require focused beams and allows scalable production of single nanopores and arrays of monodisperse nanopores for atomic-scale selectively permeable membranes.
Polymerase chain reaction (PCR) is a technique used to "amplify" small segments of DNA.
by S Li · 2020 · Cited by 2 — Graphene oxide (GO) has been suggested as an efficient assistant additive to eliminate non-specific amplification of the polymerase chain
Surface characteristics of the gate-dielectric layers in graphene field-effect transistors (FETs) critically affect the electrical properties of the devices. In this report, the effects of self-assembled monolayers (SAMs) on the electrical properties of graphene FETs were examined by using various SAM buffer layers with different end groups and alkyl chain lengths. Especially, the dipole moment of the SAMs affects the doping properties of graphene as well as field-effect mobility, hysteresis, and stability of graphene FETs. The type and magnitude of doping are dependent on the functional groups in SAMs: Electron withdrawing fluorine groups p-dope the graphene whereas electron donating amine groups n-dope the graphene. The electrical stabilities such as hysteresis and gate-bias instability are mainly governed by the magnitude of the dipole moment in SAMs.
Hexamethyldisilazane treatment resulted in graphene FETs with the highest electrical stabilities, because of the short one aliphatic alkyl chain with a negligible dipole moment. In contrast, in graphene FETs with SAMs having a strong dipole moment, electrical stabilities deteriorated by the charge trapping in SAMs.
The mRNA sequence is thus used as a template to assemble—in order—the chain of amino acids that form a protein. Figure 2: The amino acids specified by each mRNA codon. Multiple codons can code for the same amino acid. The codons are written 5' to 3', as they appear in the mRNA.
Graphene and DNA sequencing
Graphene is a material made from honeycomb sheets of carbon just one atom thick. Science journals and researchers are exhausted from trying to find new superlatives for this wondrous material: it's the lightest, strongest, thinnest, best heat and electricity conducting material ever discovered. It holds promise to revolutionize everything from computing to tennis rackets.
I
In the fields of biotechnology and medicine, it seems graphene’s thin form and malleable nature make it well suited for many possible applications such as disease and tumor detection, drug delivery, DNA sequencing and more. In DNA sequencing, a main concept is creating a graphene membrane, immerse it in conductive fluid and apply a voltage to one end so DNA can be drawn through the graphene’s miniscule pores. This method is called nanopore sequencing and it would allow DNA to be analyzed one nucleotide at a time (each nucleotide effecting the membrane differently due to its unique dimensions and electrical properties). Additional concepts involve graphene-based DNA sensors, and alternative ways of making DNA sequencing faster and more efficient.
DNA viruses have mutation rates similar to those of eukaryotic cells because, like eukaryotic DNA polymerases, their replicatory enzymes have proofreading functions. The error rate for DNA viruses has been calculated to be 10-8 to 10-11 errors per incorporated nucleotide.
Virus-induced gene mutations are probably due to insertions of fragments of viral DNA (or cDNA) into the host chromosomes; at least some of these mutations are capable of transpositions and reversions.